BRCA1_00000.gbin – empty source feature table in Genbank format
BRCA1_00000.gbin shows an empty feature-table for BRCA1_00000, with no “variation” definitions, and without a list of mRNA and CDS features.
LOCUS 17 0 bp DNA HTG 19-AUG-2022
DEFINITION Homo sapiens chromosome 17 GRCh38 partial sequence
43043295..43171245 reannotated via EnsEMBL.
ACCESSION chromosome:GRCh38:17:43043295:43171245:1
VERSION chromosome:GRCh38:17:43043295:43171245:1
KEYWORDS .
SOURCE .
ORGANISM .
.
FEATURES Location/Qualifiers
source 1..127951
/organism="Homo sapiens"
/db_xref="taxon:9606"
gene complement(1001..126951)
/gene="ENSG00000012048.24"
/locus_tag="BRCA1"
/note="BRCA1 DNA repair associated [Source:HGNC
Symbol;Acc:HGNC:1100]"
ORIGIN
//
BRCA1_hap1.gbin – source feature table in Genbank format
BRCA1_hap1.gbin shows the input “variation” features for BRCA1_hap1: where and how it differs from the reference sequence. Replicon Genetics has added “consequence” annotation taken from dbSNP
OCUS 17 0 bp DNA HTG 19-AUG-2022
DEFINITION Homo sapiens chromosome 17 GRCh38 partial sequence
43043295..43171245 reannotated via EnsEMBL.
ACCESSION chromosome:GRCh38:17:43043295:43171245:1 <--- This is the global range location for the Reference Sequence. The range is typically 2000 nucleotides longer than the FEATURE "gene" below, with 1000 additional bases at each end; the "1" refers to the sequence polarity
VERSION chromosome:GRCh38:17:43043295:43171245:1
KEYWORDS .
SOURCE .
ORGANISM .
.
COMMENT /consequence annotation by Replicon Genetics from public domain
sources Mar-2021
FEATURES Location/Qualifiers
source 1..127951 <--- This is the local range of this sequence that corresponds to the global range
/organism="Homo sapiens"
/db_xref="taxon:9606"
gene complement(1001..126951) <--- the defined region for the locus; nucleotides outside this range are "clipped" before making a Haplotype, UNLESS 'paired-end' is selected ;"complementary" shows that coding is on the opposite strand (-)
/gene="ENSG00000012048.24"
/locus_tag="BRCA1"
/note="BRCA1 DNA repair associated [Source:HGNC
Symbol;Acc:HGNC:1100]"
variation 125197 <--- This is the local-range position of the variant
/replace="T/-" <--- This is a single base deletion
/db_xref="dbSNP:rs1409504537"
/consequence="dbSNP:upstream_transcript_variant,intron_vari
ant"
variation 125198
/replace="G/A" <--- This is a single base substitution, or SNV
/db_xref="dbSNP:rs1597950091"
...
ORIGIN
//
BRCA1_hap2.gbin – source feature table in Genbank format
BRCA1_hap2.gbin shows the input “variation” features for BRCA1_hap2: where and how it differs from the reference sequence. Replicon Genetics has added “consequence”annotation taken from dbSNP
LOCUS 17 0 bp DNA HTG 19-AUG-2022
DEFINITION Homo sapiens chromosome 17 GRCh38 partial sequence
43043295..43171245 reannotated via EnsEMBL.
ACCESSION chromosome:GRCh38:17:43043295:43171245:1
VERSION chromosome:GRCh38:17:43043295:43171245:1
KEYWORDS .
SOURCE .
ORGANISM .
.
COMMENT /consequence annotation by Replicon Genetics from public domain
sources Mar-2021
FEATURES Location/Qualifiers
source 1..127951
/organism="Homo sapiens"
/db_xref="taxon:9606"
gene complement(1001..126951)
/gene="ENSG00000012048.24"
/locus_tag="BRCA1"
/note="BRCA1 DNA repair associated [Source:HGNC
Symbol;Acc:HGNC:1100]"
variation 994
/replace="T/C"
/db_xref="dbSNP:rs1411280595"
...
/replace="ATCTATCT/ATCT" <--- This is a "delins", where this variant is defined as a deletion ATCTATCT, replaced by an insert ATCT
/db_xref="dbSNP:rs776777915"
/consequence="dbSNP:intron_variant"
ORIGIN
//
BRCA1-locseq.gbin – mRNA and CDS source feature table in Genbank format
No “variation” definitions, but includes a list of mRNA and CDS features.
LOCUS 17 0 bp DNA HTG 19-AUG-2022
DEFINITION Homo sapiens chromosome 17 GRCh38 partial sequence
43043295..43171245 reannotated via EnsEMBL.
ACCESSION chromosome:GRCh38:17:43043295:43171245:1
VERSION chromosome:GRCh38:17:43043295:43171245:1
KEYWORDS .
SOURCE .
ORGANISM .
.
FEATURES Location/Qualifiers
source 1..127951
/organism="Homo sapiens"
/db_xref="taxon:9606"
gene complement(1001..126951)
/gene="ENSG00000012048.24<---
This is an Ensembl Transcript ID
/locus_tag="BRCA1"
/note="BRCA1 DNA repair associated [Source:HGNC
Symbol;Acc:HGNC:1100]"
mRNA complement(join(1001..2508,4349..4409,5827..5900,
... 63162..63239,72432..72485,80723..80821,81977..82070))
/gene="ENSG00000012048.24"
/standard_name="ENST00000357654.9"
CDS complement(join(2384..2508,4349..4409,5827..5900,
7769..7823,13758..13841,20039..20079,20580..20657,
...
BRCA1-locseq.fasta – the un-clipped, un-spliced, genomic DNA sequence of the Reference Source
>BRCA1_locseq all 127951 nucleotides from chromosome:GRCh38:17:43043295:43171245:1 AAAGGTGGCTTTGGGTCTCCATGTAGTCATTTTTAGCTGTGCAAATCTGAGTAAAATCTT ...
This sequence is the Reference Sequence for Paired-end reads.
BRCA1-locus_REF.fasta the clipped genomic DNA sequence of the Reference Source
>BRCA1-locus_REF 125951 nucleotides from 127951: End-trim of 2 regions; splice-removal of 0 regions, from BRCA1_locseq. 0 variants: 0 substitutions; 0 inserts; 0 deletions; 0 delins 1000N125951M1000N TGGAAGTGTTTGCTACCAAGTTTATTTGCAGTGTTAACAGCACAACATTTACAAAACGTA ...
If an mRNA or CDS is selected, then the name of the sequence has the format Locus–Template-{mRNA/CDS}_REF eg: BRCA1-357654-mRNA_REF . For reads other than paired-end reads this may be used as a Reference Sequence. In all cases this is simply the pre-spliced, but end-clipped, Reference Source.
BRCA1-357654-mRNA_tem.fasta – the spliced mRNA Template
This is the spliced Haplotype DNA sequence; a spliced Reference; a Template on which to merge in variants from the Variations Sources. The file name is in the format: Locus–Template-{mRNA/CDS}_tem.fasta. When the Template is Locus, the file is called BRCA1-locus_tem.fasta
>BRCA1-357654-mRNA_tem 7088 nucleotides from 127951: End-trim of 2 regions; splice-removal of 22 regions, from BRCA1_locseq. 0 variants: 0 substitutions; 0 inserts; 0 deletions; 0 delins 1000N1508M1840N61M1417N74M1868N55M5934N84M6197N41M500N78M3656N88M3232N311M3092N191M1966N127M5789N172M8368N89M402N3426M985N77M1321N46M2485N106M4241N140M606N89M1499N78M9192N54M8237N99M1155N94M45881N TGGAAGTGTTTGCTACCAAGTTTATTTGCAGTGTTAACAGCACAACATTTACAAAACGTA ...
BRCA1-locus_paired_reads.fasta example output
The first 6 entries of the reads-output file with default selections (paired-end reads), but read-length set to 20.
/1 denotes forward-reads, /2 reverse reads
>frg1_hap1 h:25988 r:25988 a:43070282 20M /2 GGTGGTAAACTTCTCAGGAT >frg1_hap1 h:26185 r:26185 a:43070479 20M /1 CTTGTAAGAATGCCCTGCCA >frg2_hap2 h:13275 r:13275 a:43057569 20M /2 CTCACGCCTGTAATCCCAGG >frg2_hap2 h:13471 r:13471 a:43057765 20M /1 CTCCCGGGTTCACGCCATTC >frg3_hap2 h:90786 r:90786 a:43135080 20M /1 CCACGTGTCTTGCTCTGGCC >frg3_hap2 h:90985 r:90985 a:43135279 20M /2 CCTGCAGGCCTGCGGATCGG
BRCA1-locus_paired_reads.fastq example output
BRCA1-locus_paired_reads.fastq contains the same 6 reads as the above BRCA1-locus_paired_reads.fasta file, but with a quality line for each read
@frg1_hap1 h:25988 r:25988 a:43070282 20M /2 GGTGGTAAACTTCTCAGGAT + 35BBGGSK8J3J>1K6>S52 @frg1_hap1 h:26185 r:26185 a:43070479 20M /1 CTTGTAAGAATGCCCTGCCA + 9?SG<G=N8GE125>EIO:2 @frg2_hap2 h:13275 r:13275 a:43057569 20M /2 CTCACGCCTGTAATCCCAGG + =EQLA1I578DRO<L725QA @frg2_hap2 h:13471 r:13471 a:43057765 20M /1 CTCCCGGGTTCACGCCATTC + <I1@IK48SRGQ>9:I5E>0 @frg3_hap2 h:90786 r:90786 a:43135080 20M /1 CCACGTGTCTTGCTCTGGCC + 9QDOK7IBSN@8AL9DOK3J @frg3_hap2 h:90985 r:90985 a:43135279 20M /2 CCTGCAGGCCTGCGGATCGG + Q4@11>4FAR9O0CHF23C@
BRCA1-locus_hap2.gbout – source feature table in Genbank format
BRCA1-locus_hap2.gbout shows the absolute genomic position for any variation features seen in BRCA1_hap2.gbin
The content of this file is different depending on whether Paired reads is selected (no trimmed ends) or not-selected ( trimmed ends are shown as /replace=”N/-“, equivalent to deletions). Any variants located in the trimmed regions defined in the .gbin file do not appear in the .gbout file
LOCUS 17 0 bp DNA HTG 19-AUG-2022
DEFINITION Homo sapiens chromosome 17 GRCh38 partial sequence
43043295..43171245 reannotated via EnsEMBL.
ACCESSION chromosome:GRCh38:17:43043295:43171245:1
VERSION chromosome:GRCh38:17:43043295:43171245:1
KEYWORDS .
SOURCE .
ORGANISM .
.
FEATURES Location/Qualifiers
source 1..127951
/organism="Homo sapiens"
/db_xref="taxon:9606"
gene complement(1001..126951)
/gene="ENSG00000012048.24"
/locus_tag="BRCA1"
/note="BRCA1 DNA repair associated [Source:HGNC
Symbol;Acc:HGNC:1100]"
<--- only shown in the trimmed version ...
variation 126952..127951
/replace="N/-"
/db_xref="gap:3-prime downstream trim"
/global_range="GRCh38:17:43170246:43171245:1"
... only shown in the trimmed version --->
variation 124950..124957
/replace="ATCTATCT/ATCT"
/db_xref="dbSNP:rs776777915"
/consequence="dbSNP:intron_variant"
/global_range="GRCh38:17:43168244:43168251:1"
variation 124942
/replace="G/A"
/db_xref="dbSNP:rs531592442"
/consequence="dbSNP:intron_variant"
/global_range="GRCh38:17:43168236:43168236:1"
variation 124934
/replace="C/T"
/db_xref="dbSNP:rs887555188"
/consequence="dbSNP:intron_variant"
/global_range="GRCh38:17:43168228:43168228:1"
variation 124933
/replace="A/C"
/db_xref="dbSNP:rs1300100629"
/consequence="dbSNP:intron_variant"
/global_range="GRCh38:17:43168227:43168227:1"
variation 124927
/replace="T/C"
/db_xref="dbSNP:rs1321005885"
/consequence="dbSNP:intron_variant"
/global_range="GRCh38:17:43168221:43168221:1"
variation 82002
/replace="C/G"
/db_xref="dbSNP:rs1057521869"
/consequence="dbSNP:genic_upstream_transcript_variant,non_c
oding_transcript_variant"
/consequence="dbSNP:upstream_transcript_variant,5_prime_UTR
_variant"
/global_range="GRCh38:17:43125296:43125296:1"
variation 80822
/replace="C/T"
/db_xref="dbSNP:rs569074958"
/consequence="dbSNP:genic_upstream_transcript_variant"
/consequence="dbSNP:upstream_transcript_variant,splice_acce
ptor_variant"
/global_range="GRCh38:17:43124116:43124116:1"
variation 80818
/replace="T/C"
/db_xref="dbSNP:rs777262055"
/consequence="dbSNP:upstream_transcript_variant,5_prime_UTR
_variant"
/consequence="dbSNP:non_coding_transcript_variant"
/global_range="GRCh38:17:43124112:43124112:1"
variation 72490
/replace="C/T"
/db_xref="dbSNP:rs1555599296"
/consequence="dbSNP:intron_variant"
/global_range="GRCh38:17:43115784:43115784:1"
variation 72482
/replace="C/T"
/db_xref="dbSNP:rs1555599278"
/consequence="dbSNP:intron_variant,non_coding_transcript_va
riant"
/consequence="dbSNP:synonymous_variant,coding_sequence_vari
ant"
/global_range="GRCh38:17:43115776:43115776:1"
variation 4402
/replace="T/G"
/db_xref="dbSNP:rs397509281"
/consequence="dbSNP:non_coding_transcript_variant,synonymou
s_variant"
/consequence="dbSNP:coding_sequence_variant,missense_varian
t"
/global_range="GRCh38:17:43047696:43047696:1"
variation 4332
/replace="A/T"
/db_xref="dbSNP:rs1267019068"
/consequence="dbSNP:intron_variant"
/global_range="GRCh38:17:43047626:43047626:1"
variation 1102
/replace="G/T"
/db_xref="dbSNP:rs1304626969"
variation 1102
/replace="G/T"
/db_xref="dbSNP:rs1304626969"
/consequence="dbSNP:non_coding_transcript_variant,3_prime_U
TR_variant"
/global_range="GRCh38:17:43044396:43044396:1"
<--- only shown in the trimmed version ...
variation 1..1000
/replace="N/-"
/db_xref="gap:5-prime upstream trim"
/global_range="GRCh38:17:43043295:43044294:1"
/consequence="dbSNP:non_coding_transcript_variant,3_prime_U
TR_variant"
/global_range="GRCh38:17:43044396:43044396:1"
variation 994
/replace="T/C"
/db_xref="dbSNP:rs1411280595"
/consequence="dbSNP:downstream_transcript_variant"
/global_range="GRCh38:17:43044288:43044288:1"
... only shown in the trimmed version --->
ORIGIN
//
BRCA1-357654-mRNA_hap2.gbout source feature table in Genbank
BRCA1-357654-mRNA_hap2.gbout shows the absolute genomic position for clipped sections and introns as deletions, plus all variation features seen in BRCA1_hap2.gbin that have been retained in exons. Any variant features that are defined within introns have been eliminated; any variant features that cross intron-exon boundaries would be clipped (not shown here).
This version shows the gaps that replace intron sequence, and excludes any “variation” features that were present within those gaps in the above version for genomic BRCA1_hap2.gbout
LOCUS 17 0 bp DNA HTG 19-AUG-2022
DEFINITION Homo sapiens chromosome 17 GRCh38 partial sequence
43043295..43171245 reannotated via EnsEMBL.
ACCESSION chromosome:GRCh38:17:43043295:43171245:1
VERSION chromosome:GRCh38:17:43043295:43171245:1
KEYWORDS .
SOURCE .
ORGANISM .
.
FEATURES Location/Qualifiers
source 1..127951
/organism="Homo sapiens"
/db_xref="taxon:9606"
gene complement(1001..126951)
/gene="ENSG00000012048.24"
/locus_tag="BRCA1"
/note="BRCA1 DNA repair associated [Source:HGNC
Symbol;Acc:HGNC:1100]"
variation 82071..127951
/replace="N/-"
/db_xref="gap:3-prime downstream trim"
/global_range="GRCh38:17:43125365:43171245:1 <--- This is the global range location for the 3' trim in the Reference Source NB: all variants up to this point defined in BRCA1_hap2.gbout are absent
variation 82002
/replace="C/G"
/db_xref="dbSNP:rs1057521869"
/consequence="dbSNP:genic_upstream_transcript_variant,non_c
oding_transcript_variant"
/consequence="dbSNP:upstream_transcript_variant,5_prime_UTR
_variant"
/global_range="GRCh38:17:43125296:43125296:1" <--- This is the global location for this SNP
variation 80822..81976
/replace="N/-"
/db_xref="gap:Intron 22-23"
/global_range="GRCh38:17:43124116:43125270:1"
variation 80818
/replace="T/C"
/db_xref="dbSNP:rs777262055"
/consequence="dbSNP:upstream_transcript_variant,5_prime_UTR
_variant"
/consequence="dbSNP:non_coding_transcript_variant"
/global_range="GRCh38:17:43124112:43124112:1"
variation 72486..80722
/replace="N/-"
/db_xref="gap:Intron 21-22"
/global_range="GRCh38:17:43115780:43124016:1" <--- This is the global range location for the intron between exons 21 and 22
variation 72482
/replace="C/T"
/db_xref="dbSNP:rs1555599278"
/consequence="dbSNP:intron_variant,non_coding_transcript_va
riant" /consequence="dbSNP:synonymous_variant,coding_sequence_vari
ant"
/global_range="GRCh38:17:43115776:43115776:1"
variation 63240..72431
/replace="N/-"
/db_xref="gap:Intron 20-21"
/global_range="GRCh38:17:43106534:43115725:1"
variation 61663..63161
/replace="N/-"
/db_xref="gap:Intron 19-20"
/global_range="GRCh38:17:43104957:43106455:1"
variation 60968..61573
/replace="N/-"
/db_xref="gap:Intron 18-19"
/global_range="GRCh38:17:43104262:43104867:1"
variation 56587..60827
/replace="N/-"
/db_xref="gap:Intron 17-18"
/global_range="GRCh38:17:43099881:43104121:1"
variation 53996..56480
/replace="N/-"
/db_xref="gap:Intron 16-17"
/global_range="GRCh38:17:43097290:43099774:1"
variation 52629..53949
/replace="N/-"
/db_xref="gap:Intron 15-16"
/global_range="GRCh38:17:43095923:43097243:1"
variation 51567..52551
/replace="N/-"
/db_xref="gap:Intron 14-15"
/global_range="GRCh38:17:43094861:43095845:1"
variation 47739..48140
/replace="N/-"
/db_xref="gap:Intron 13-14"
/global_range="GRCh38:17:43091033:43091434:1"
variation 39282..47649
/replace="N/-"
/db_xref="gap:Intron 12-13"
/global_range="GRCh38:17:43082576:43090943:1"
variation 33321..39109
/replace="N/-"
/db_xref="gap:Intron 11-12"
/global_range="GRCh38:17:43076615:43082403:1"
variation 31228..33193
/replace="N/-"
/db_xref="gap:Intron 10-11"
/global_range="GRCh38:17:43074522:43076487:1"
variation 27945..31036
/replace="N/-"
/db_xref="gap:Intron 9-10"
/global_range="GRCh38:17:43071239:43074330:1"
variation 24402..27633
/replace="N/-"
/db_xref="gap:Intron 8-9"
/global_range="GRCh38:17:43067696:43070927:1"
variation 20658..24313
/replace="N/-"
/db_xref="gap:Intron 7-8"
/global_range="GRCh38:17:43063952:43067607:1"
variation 20080..20579
/replace="N/-"
/db_xref="gap:Intron 6-7"
/global_range="GRCh38:17:43063374:43063873:1"
variation 13842..20038
/replace="N/-"
/db_xref="gap:Intron 5-6"
/global_range="GRCh38:17:43057136:43063332:1"
variation 7824..13757
/replace="N/-"
/db_xref="gap:Intron 4-5"
/global_range="GRCh38:17:43051118:43057051:1"
variation 5901..7768
/replace="N/-"
/db_xref="gap:Intron 3-4"
/global_range="GRCh38:17:43049195:43051062:1"
variation 4410..5826
/replace="N/-"
/db_xref="gap:Intron 2-3"
/global_range="GRCh38:17:43047704:43049120:1"
variation 4402
/replace="T/G"
/db_xref="dbSNP:rs397509281"
/consequence="dbSNP:non_coding_transcript_variant,synonymou
s_variant" /consequence="dbSNP:coding_sequence_variant,missense_varian
t"
/global_range="GRCh38:17:43047696:43047696:1"
NB: dbSNP rs1267019068 at local position 4332 is not present in this file because it sits within intron 1-2
variation 2509..4348
/replace="N/-"
/db_xref="gap:Intron 1-2"
/global_range="GRCh38:17:43045803:43047642:1"
variation 1102
/replace="G/T"
/db_xref="dbSNP:rs1304626969"
/consequence="dbSNP:non_coding_transcript_variant,3_prime_U
TR_variant"
/global_range="GRCh38:17:43044396:43044396:1"
variation 1..1000
/replace="N/-"
/db_xref="gap:5-prime upstream trim"
/global_range="GRCh38:17:43043295:43044294:1"
ORIGIN
//
BRCA1-gene_paired_reads.sam
This is the SAM format file showing how paired-end synthetic reads would align against the reference sequence in a perfect solution. Please note that no alignment algorithm has been run. It is only possible to produce this file because all reads are synthetic, and the application has tracked the start and end positions of the reads from the original Reference Source.
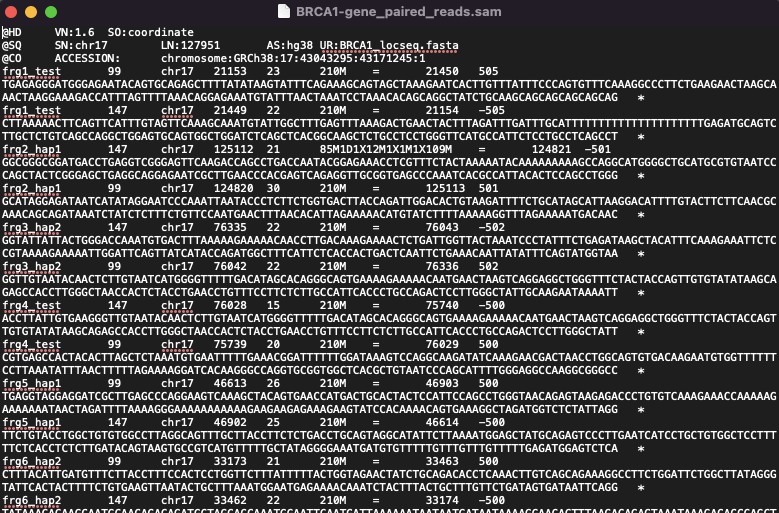
The SAM file can be processed for display by IGV using samtools:
% samtools view -@ n -Sb -o BRCA1-gene_paired_reads.bam BRCA1-gene_paired_reads.sam
% samtools sort -O bam -o BRCA1-gene_paired_reads_alignment.bam BRCA1-gene_paired_reads.bam
% samtools index BRCA1-gene_paired_reads_alignment.bam
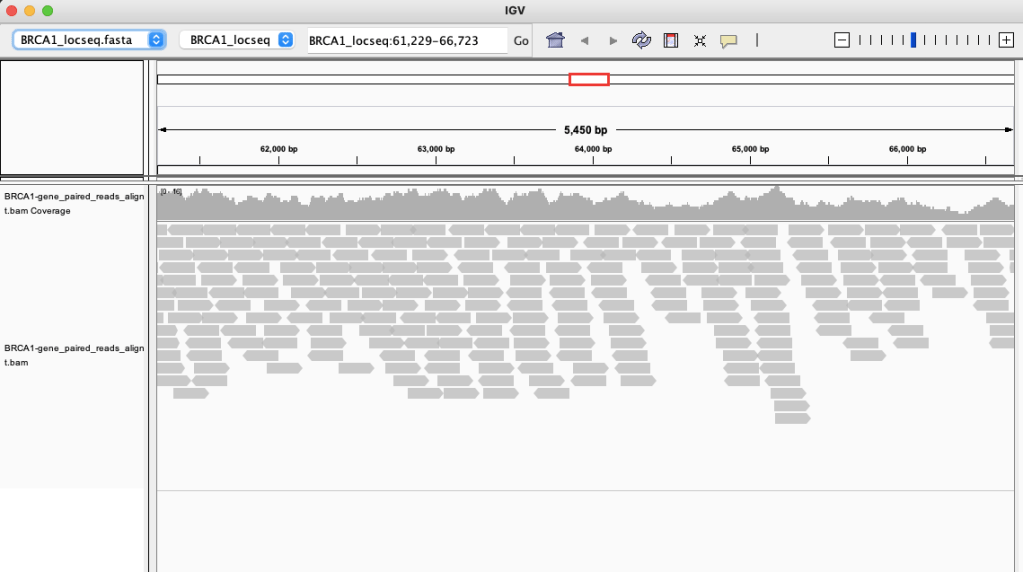
In IGV, after “Load Genome from file”, and selecting BRCA1_locseq.fasta (use “Save Reference Haplotype”) as the Genome; then loading BRCA1-gene_paired_reads_alignment.bam into IGV, you get this:
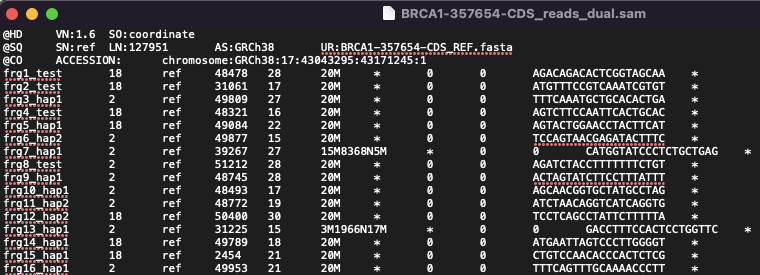
BRCA1-357654-CDS_reads_dual.sam
This is the SAM format file showing how the synthetic reads would align against the reference sequence in a perfect solution. Please note that no alignment algorithm has been run. It is only possible to produce this file because all reads are synthetic, and the application has tracked the start and end positions of the reads from the original Reference Source.
BRCA1-locus_000_paired_readme
Example contents of BRCA1-locus_000_paired_readme – contents will differ depending on the options selected
This readme file BRCA1-locus_000_paired_readme is written by Program RG_exploder_main_23_7.py 25-Aug-2022 starting on Wed Aug 31 16:14:13 2022 Read in conjunction with BRCA1-locus_001_paired_journal Program input files: BRCA1_locseq.gb - 'Reference Source' BRCA1_hap1.gb - 'Variations Source' BRCA1_hap2.gb - 'Variations Source' Program output metadata files: BRCA1-locus_000_readme - This file BRCA1-locus_001_journal - Journal file documenting runtime messages & metadata including 'Reference Source' headers, program parameters BRCA1-locus_001_journal.htm - Journal file documenting runtime messages & metadata including 'Reference Source' headers, program parameters (html version) BRCA1-locus_002_paired_config.txt - contains configuration data for this run BRCA1_locseq.gbin - Feature definitions from the 'Reference Source' BRCA1_locseq BRCA1_hap1.gbin - Initial feature list for Variations Source 'hap1' BRCA1-locus_hap1.gbout - Feature definitions for Variations Source 'hap1' absolute positions added BRCA1_hap2.gbin - Initial feature list for Variations Source 'hap2' BRCA1-locus_hap2.gbout - Feature definitions for Variations Source 'hap2' absolute positions added Program output sequence files: BRCA1_locseq.fasta - FASTA file of the un-modified 'Reference Source' BRCA1_locseq sequence BRCA1-locus_REF.fasta - FASTA file used to select the first in a pair of paired-ends BRCA1-locus_var.fasta - FASTA file of all Haplotype Definition with frequency >0: hap1, hap2; BRCA1-locus_paired_reads.fasta - FASTA sequence reads from all Haplotype Definition with frequency >0: hap1, hap2 BRCA1-locus_paired_reads.fastq - FASTQ sequence reads from BRCA1-locus_paired_reads.fasta with a random quality score between 15-50 at each base BRCA1-locus_paired_reads.sam - SAM file of all the sequence reads from BRCA1-locus_paired_reads.fasta Ending RG_exploder_main_23_7.py at Wed Aug 31 16:14:27 2022 Total time taken:13.148000001907349 Copyright © Replicon Genetics 2021, 2022. All rights reserved.
BRCA1-locus_001_paired_journal
Example contents of BRCA1-locus_001_paired_journal – contents will differ depending on the options selected
This journal file BRCA1-locus_001_paired_journal is created by RG_exploder_main_23_7.py 25-Aug-2022 starting on Wed Aug 31 16:14:13 2022
Read in conjunction with BRCA1-locus_000_paired_readme
User ID:Public
Data set:Open Access GRCh38; August 2022
Selected Locus: BRCA1; Selected Template: Locus; Selected CDS only: False
If this is the last line, then something has gone wrong reading the source files
Reading 'Reference Source' file BRCA1_locseq.gb
BRCA1_locseq 'Reference Source' Range defined as: GRCh38:17:43043295:43171245:1
BRCA1_locseq 'Reference Source' correctly includes 0 variant features
MaxVarPos set to full sequence length of 127951 bases from 'Reference Source' BRCA1_locseq
Feature definitions for 'Reference Source' BRCA1_locseq saved as BRCA1_locseq.gbin
Writing BRCA1_locseq.gbin
Searching feature table to set Reference Template
Locus BRCA1 matches gene id ENSG00000012048.24 from BRCA1_locseq Range: 1001 - 126951 ; global : 43044295 - 43170245; length: 125951 bases
With option '(Exome) Extension'=0, Template Range from BRCA1_locseq also: 1001 - 126951 ; global : 43044295 - 43170245; length: 125951 bases
Template BRCA1-locus has 125951 bases; 2 spliced-out regions compared to 'Reference Source' BRCA1_locseq
No splicing of exon boundaries because 'Template' is set to 'Locus'
Exact match between Source Ranges for BRCA1_REF and BRCA1_locseq
BRCA1-locus_REF length: 125951 bases. End-trim of 2 regions; splice-removal of 0 regions, from BRCA1_locseq. 0 variants: 0 substitutions; 0 inserts; 0 deletions; 0 delins
BRCA1-locus_REF CIGAR(wrt BRCA1_locseq): 1000N125951M1000N
Writing BRCA1-locus_REF.fasta
BRCA1-locus_REF, Length: 125951 bases, is the ** Trimmed BRCA1_locseq.fasta ** for the paired reads used to select the first in a pair of paired-ends. The second may be partly or fully outside this sequence, but fully within the Reference Sequence.
Writing BRCA1_locseq.fasta
BRCA1_locseq Location: GRCh38:17:43043295:43171245:1; length: all 127951 of 127951 bases
BRCA1_locseq.fasta, Length: 127951 bases, is the ** Reference Sequence ** for the paired-end reads in BRCA1-locus_paired_reads.fasta, BRCA1-locus_paired_reads.fastq and BRCA1-locus_paired-reads.sam
Reading Variations Source (feature) files...
Reading 'Variations Source' file BRCA1_hap1.gb
Writing BRCA1_hap1.gbin
Compatible GRCh build, chromosome and polarity: GRCh38:17:1
Matching ranges: Reference Source_range: 43043295:43171245; Variations Source_range: 43043295:43171245
Not splicing Locus
Writing BRCA1-locus_hap1.gbout
Exact match between Source Ranges for BRCA1_hap1 and BRCA1_locseq
BRCA1-locus_hap1 length: 127950 bases. End-trim of 0 regions; splice-removal of 0 regions, from BRCA1_locseq. 4 variants: 3 substitutions; 0 inserts; 1 deletions; 0 delins
BRCA1-locus_hap1 CIGAR(wrt BRCA1_locseq): 125196M1D1X12M1X1M1X2738M
Writing BRCA1-locus_hap1 FASTA to BRCA1-locus_var.fasta
Reading 'Variations Source' file BRCA1_hap2.gb
Writing BRCA1_hap2.gbin
Compatible GRCh build, chromosome and polarity: GRCh38:17:1
Matching ranges: Reference Source_range: 43043295:43171245; Variations Source_range: 43043295:43171245
Not splicing Locus
Writing BRCA1-locus_hap2.gbout
Exact match between Source Ranges for BRCA1_hap2 and BRCA1_locseq
BRCA1-locus_hap2 length: 127947 bases. End-trim of 0 regions; splice-removal of 0 regions, from BRCA1_locseq. 14 variants: 13 substitutions; 0 inserts; 0 deletions; 1 delins
BRCA1-locus_hap2 CIGAR(wrt BRCA1_locseq): 993M1X107M1X3229M1X69M1X68079M1X7M1X8327M1X3M1X1179M1X42924M1X5M2X7M1X7M8D4I2994M
Writing BRCA1-locus_hap2 FASTA to BRCA1-locus_var.fasta
Processing 2 Haplotype Definitions with frequency >0 : hap1, hap2
Relative frequency values : [50,50]
Normalised proportion values: [500,500]
Normalised ratios: [1.0,1.0]
Writing FASTA reads to BRCA1-locus_paired_reads.fasta
Writing FASTQ reads to BRCA1-locus_paired-reads.fastq
FASTQ quality range 15 to 50
FASTQ quality range has randomly-assigned quality values in each read
Writing reads in SAM format to BRCA1-locus_paired_reads.sam
For a 'Depth of cover' target value of 3, based on reference length 127951:
Generated 19194 reads of length 20 bases at random starting positions within 2 Haplotype Definitions: hap1, hap2
source(count):
hap1(9590),hap2(9604)
source(count-ratio):
hap1(1.0),hap2(1.0)
source(length):
hap1(127950),hap2(127947)
source('Depth of cover'=count*20/length):
hap1(1.5),hap2(1.5)
Read length=20
Total number of reads=19194
0 sections from the Reference Sequence are spliced out
Saved reads created as single strand: forward only
Ending RG_exploder_main_23_7.py at Wed Aug 31 16:14:27 2022
Total time taken:13.148000001907349
Copyright © Replicon Genetics 2021, 2022. All rights reserved.
BRCA1-locus_002_paired_config.txt
Example contents of BRCA1-locus_002_paired_config.txt- contents will differ depending on the options selected
{
"custom_stringconstants": {
"DatasetIDText": "Open Access GRCh38; August 2022",
"CustomerIDText": "Public",
"GUI_ConfigText": "Configuration at Wed Aug 31 16:14:13 2022"
},
"bio_parameters": {
"target_locus": {
"label": "Locus",
"value": "BRCA1"
},
"target_transcript_name": {
"label": "Template",
"value": "Locus"
},
"target_transcript_id": {
"value": ""
},
"target_build_variant": {
"is_get_ref": false,
"is_save_var": false,
"is_get_muttranscripts": false,
"is_join_complement": false,
"mRNA_join": "",
"CDS_join": "",
"mrnapos_lookup": "hidden",
"transcript_view": "",
"abs_offset": 0,
"ref_strand": 1,
"max_seqlength": 0,
"ref_label": "Reference Sequence",
"var_label": "Variant Sequence",
"var_name_label": "Variant Name",
"var_name": "",
"ref_start": 0,
"ref_end": 0,
"ref_subseq": "",
"ref_viewstring": "",
"var_subseq": "",
"AddVars": []
},
"is_CDS": {
"label": "CDS only",
"value": false
},
"mutfreqs": {
"00000": 0,
"hap1": 50,
"hap2": 50,
"test": 0
},
"Fraglen": {
"label": "Read length",
"value": 20,
"min": 4,
"max": 2000
},
"Fragdepth": {
"label": "Depth of cover",
"value": 3,
"min": 1,
"max": 500
},
"Exome_extend": {
"label": "(Exome) Extension",
"value": 0,
"min": 0,
"max": 50
},
"is_flip_strand": {
"label": "Flip polarity",
"value": false
},
"is_frg_paired_end": {
"label": "Paired-end",
"value": true
},
"is_duplex": {
"label": "Dual-strand",
"value": false
},
"is_simplex": {
"label": "Single-strand",
"value": null
},
"is_fasta_out": {
"label": "Reads in FASTA format",
"value": true
},
"is_onefrag_out": {
"label": "- Each possible read",
"value": false
},
"is_muts_only": {
"label": "- Variant reads only",
"value": false
},
"is_frg_label": {
"label": "- Annotate source positions...",
"value": true
},
"is_use_absolute": {
"label": "- ... plus absolute position",
"value": true
},
"is_fastacigar_out": {
"label": "- CIGAR annotation",
"value": true
},
"is_vars_to_lower": {
"label": "- Substitutions in lower case",
"value": false
},
"is_journal_subs": {
"label": "- Journal the substitutions",
"value": false
},
"is_fastq_out": {
"label": "Reads in FASTQ format",
"value": true
},
"Qualmin": {
"label": "- FASTQ quality min",
"value": 15,
"min": 0,
"max": 93
},
"Qualmax": {
"label": "- FASTQ quality max",
"value": 50,
"min": 0,
"max": 93
},
"is_write_ref_fasta": {
"label": "Save Reference Sequences",
"value": true
},
"is_mut_out": {
"label": "Save Haplotype Sequences",
"value": true
},
"is_write_ref_ingb": {
"label": "Save Source Features",
"value": true
},
"is_sam_out": {
"label": "Reads in SAM format",
"value": true
},
"gauss_mean": {
"label": "Mean insert size",
"value": 200,
"min": 100,
"max": 400
},
"gauss_SD": {
"label": "SD insert size",
"value": 2,
"min": 0,
"max": 20
}
},
"Reference_sequences": {
"BRCA1": {
"Release": "Ensembl Release 105 (Dec 2021)",
"Retrieval_date": "19-AUG-2022",
"Region": "GRCh38:17:43043295:43171245:1",
"Locus_range": "1001:126951",
"is_join_complement": true,
"LRG_id": "292",
"Ensembl_id": "ENSG00000012048.24",
"mRNA": {
"BRCA1-357654(MANE_Select)": "ENST00000357654.9",
"BRCA1-352993": "ENST00000352993.7",
"BRCA1-354071": "ENST00000354071.7",
"BRCA1-412061": "ENST00000412061.3",
"BRCA1-461221": "ENST00000461221.5",
"BRCA1-461574": "ENST00000461574.1",
"BRCA1-461798": "ENST00000461798.5",
"BRCA1-468300": "ENST00000468300.5",
"BRCA1-470026": "ENST00000470026.5",
"BRCA1-471181": "ENST00000471181.7",
"BRCA1-473961": "ENST00000473961.5",
"BRCA1-476777": "ENST00000476777.5",
"BRCA1-477152": "ENST00000477152.5",
"BRCA1-478531": "ENST00000478531.5",
"BRCA1-484087": "ENST00000484087.6",
"BRCA1-489037": "ENST00000489037.1",
"BRCA1-491747": "ENST00000491747.6",
"BRCA1-492859": "ENST00000492859.5",
"BRCA1-493795": "ENST00000493795.5",
"BRCA1-493919": "ENST00000493919.5",
"BRCA1-494123": "ENST00000494123.5",
"BRCA1-497488": "ENST00000497488.1",
"BRCA1-586385": "ENST00000586385.5",
"BRCA1-591534": "ENST00000591534.5",
"BRCA1-591849": "ENST00000591849.5",
"BRCA1-618469": "ENST00000618469.1",
"BRCA1-634433": "ENST00000634433.1",
"BRCA1-642945": "ENST00000642945.1",
"BRCA1-644379": "ENST00000644379.1",
"BRCA1-644555": "ENST00000644555.1",
"BRCA1-652672": "ENST00000652672.1",
"BRCA1-700182": "ENST00000700182.1",
"BRCA1-700183": "ENST00000700183.1"
},
"mRNA_join": "",
"CDS_join": "",
"MANE_Select": {
"version": "v0.95",
"mRNA": "BRCA1-357654(MANE_Select)"
}
}
}
}